Research Areas
Dynamics of multi-protein complexes at the single-molecule level
We want to obtain a real time picture of how components of a multi-protein complex interact to perform complex regulated tasks, in particular to see how a protein system might be more than the sum of its components. In living organisms many proteins work in complexes to form multicomponent protein machines and to regulate cellular processes. The function of such multicomponent machines is usually addressed by dividing them into a collection of two state systems at equilibrium. Many molecular machines, like e.g. the heat shock protein Hsp90, work in large complexes with multiple states out of equilibrium by utilizing the energy of ATP hydrolysis.
These studies impact the understanding of the Hsp90 machinery as well as general principles of multi-component protein systems, which is the basis for understanding cellular processes.
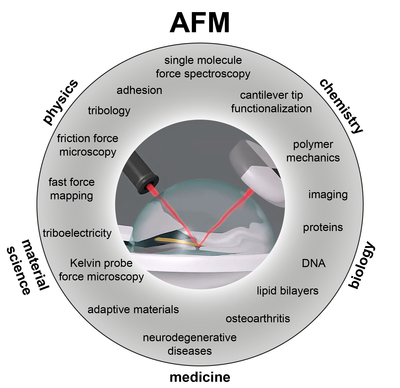
Atomic Force Microscopy
Polymers at interfaces play an important role in (bio)technology, but a fundamental understanding of their interaction is still lacking. We develop AFM-based methods to probe single polymer adhesion and friction at interfaces. Experiments of this kind will help in gaining a fundamental understanding of adhesion and friction properties of multicomponent systems. Furthemore, we extend our methods to analyze the mechnics of adaptive material systems, the charge separation properties of troboelectric materials and the elasticity and structure of cartilage for the understanding of osteoarthristis.
AFM website: Balzer lab
Publications
Methods
Software
Contact
FRET: thorsten.hugel@physchem.uni-freiburg.de
AFM: bizan.balzer@physchem.uni-freiburg.de
Biochemistry: bianca.hermann@physchem.uni-freiburg.de